Embedding High-dimensional Bayesian Optimization via Generative Modeling - Parameter Personalization of Cardiac Electrophysiological Models
Abstract
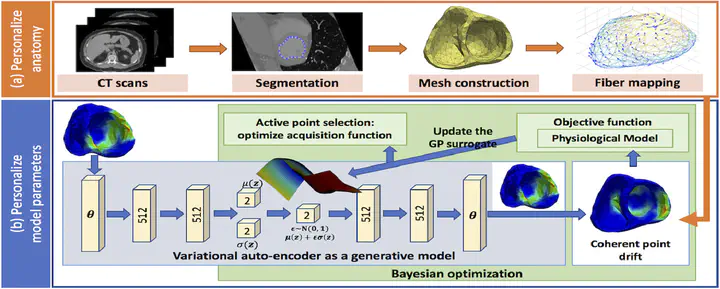
The estimation of patient-specific tissue properties in the form of model parameters is important for personalized physiological models. Because tissue properties are spatially varying across the underlying geometrical model, it presents a significant challenge of high-dimensional (HD) optimization at the presence of limited measurement data. A common solution to reduce the dimension of the parameter space is to explicitly partition the geometrical mesh. In this paper, we present a novel concept that uses a generative variational auto-encoder (VAE) to embed HD Bayesian optimization into a low-dimensional (LD) latent space that represents the generative code of HD parameters. We further utilize VAE-encoded knowledge about the generative code to guide the exploration of the search space. The presented method is applied to estimating tissue excitability in a cardiac electrophysiological model in a range of synthetic and real-data experiments, through which we demonstrate its improved accuracy and substantially reduced computational cost in comparison to existing methods that rely on geometry-based reduction of the HD parameter space.